EVENT
【Seminar Report】WPI-Bio2Q Open Seminar
January 8, 2026

Dr. Masahiro Kanai
Credits: WPI-Bio2Q

Group Photo
Credits: WPI-Bio2Q
Poster
Credits: WPI-Bio2Q






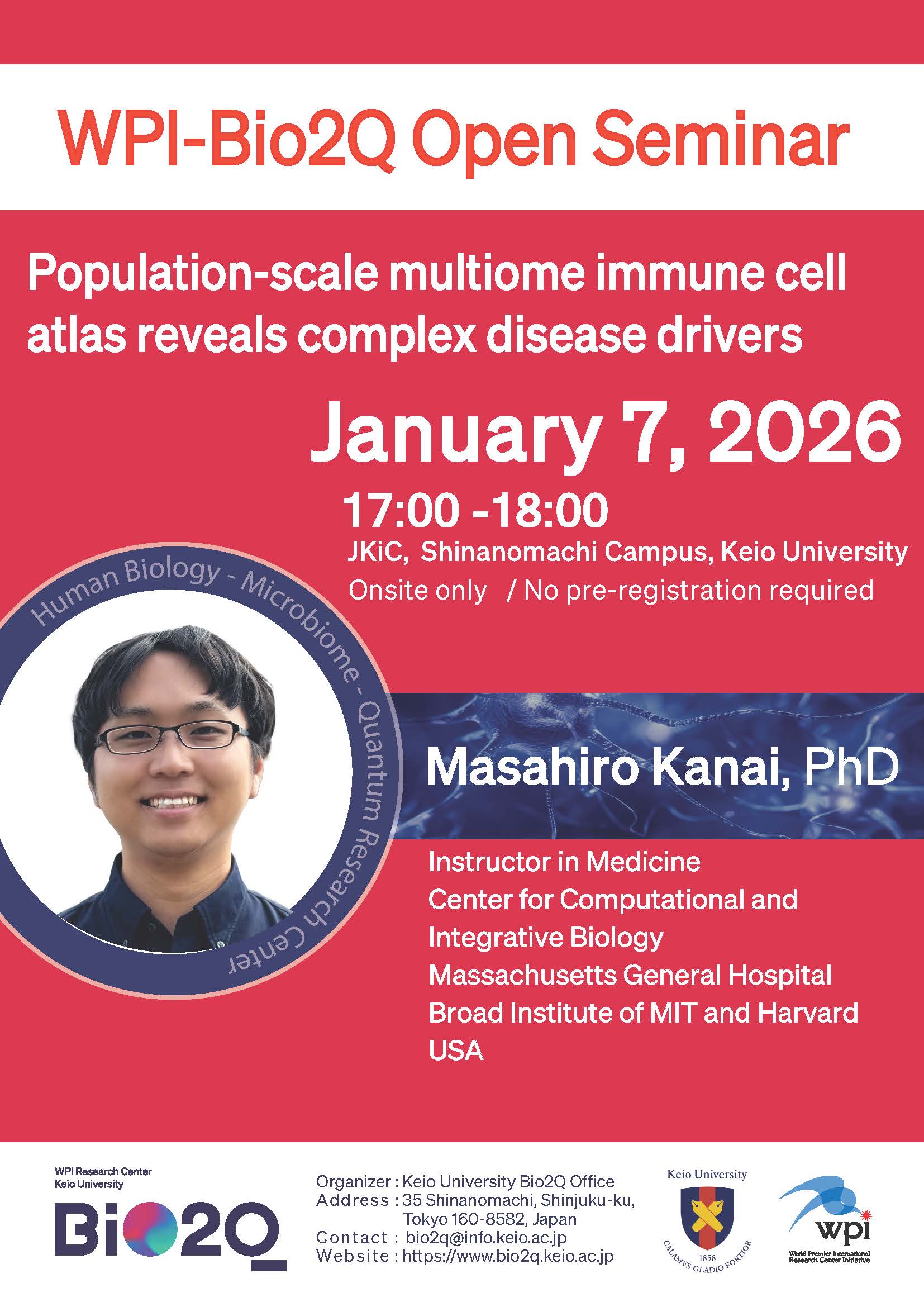
Keio University Human Biology-Microbiome-Quantum Research Center (WPI-Bio2Q) held a seminar as follows.
17:00 -18:00 January 7, 2026
Venue: JKiC1F, Shinanomachi Campus, Keio University
Speaker:
Masahiro Kanai, PhD
Instructor in Medicine, Center for Computational and Integrative Biology
Massachusetts General Hospital
Broad Institute of MIT and Harvard
USA
Title: “Population-scale multiome immune cell atlas reveals complex disease drivers”
Abstract: “Most genetic variants associated with complex diseases lie in non-coding regions, yet mechanistic insights have been limited by the lack of an empirical framework for characterizing the molecular consequences of regulatory variation. Single-cell profiling of molecular quantitative trait loci (QTL) can connect variants to gene regulation, but prior studies lacked the sample size to detect variants at disease-relevant genes and the simultaneous measurements across regulatory layers needed to trace complete mechanisms from chromatin state to gene expression. Here we show that population-scale simultaneous profiling of chromatin accessibility and gene expression across immune cell types reveals multi-layered regulatory pathways connecting genetic variants to disease. We generated paired single-nucleus ATAC-seq and RNA-seq profiles from 10 million peripheral blood mononuclear cells across 1,108 Finnish individuals, identifying 51,083 cis-eQTLs for 20,829 genes, 338,100 cis-caQTLs for 210,584 peaks, 119,094 fine-mapped variants, and 496,488 enhancer–gene links. Systematic classification of regulatory mechanisms revealed that variants with complete chromatin-to-expression cascades show twice the disease colocalization of chromatin-only effects, establishing a hierarchy where mechanistic cascade predicts disease relevance. Analysis of evolutionarily constrained genes revealed multi-layered regulatory buffering where chromatin accessibility changes occur with normal effect sizes, but transmission to gene expression is attenuated through systematically weaker enhancer–gene links, reconciling why disease variants preferentially target these genes despite apparent eQTL depletion. We incorporated base editing to experimentally validate causal variants and mechanisms at Finnish-enriched disease loci such as TNRC18. This resource provides testable mechanistic hypotheses for over half of immune disease associations.”
More Bio2Q News
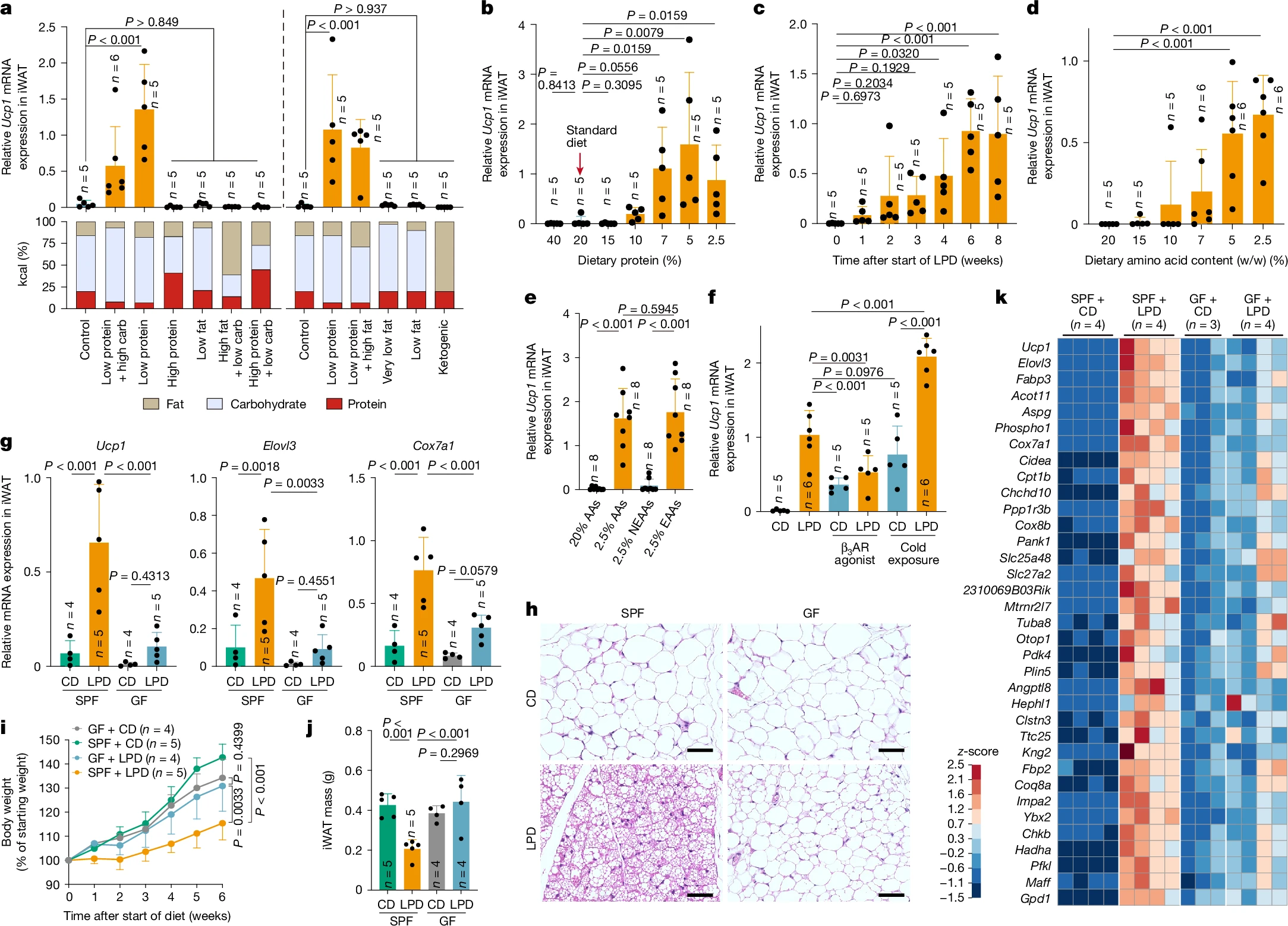
[Publication] Microbiota-mediated Induction of Beige Adipocytes in Response to D...
A research group led by Bio2Q Center Director Professor Kenya Honda has revealed how a low-protein diet induces energy-burning beige adipocy...
New Member - Dr. Leonard Dubois
Introducing new member of Bio2Q We are excited to welcome Dr. Leonard Dubois as a Postdoctoral Fellow at Bio-1 Core of Bio2Q. "Hello, I ...
【Event Report】WPI Young Researchers Forum
Keio University Human Biology-Microbiome-Quantum Research Center (WPI-Bio2Q) participated in the WPI Young Researchers Forum. WPI Young R...
Engaging Discussion on Cutting-Edge Research with Dr. Magdalena Skipper, Editor-...
On February 20, Dr. Magdalena Skipper, Editor-in-Chief of the prestigious scientific journal Nature, visited CRIK Shinanomachi together wit...
New Member - Dr. Jung Hyun Im
Introducing new member of Bio2Q We are excited to welcome Dr. Jung Hyun Im as a Postdoctoral Fellow at Bio-1 Core of Bio2Q. "Hello, ever...